Element Biosciences is launching their AVITI sequencing system today in a blitz of events. At February’s end they flew me out to visit their San Diego facility and gave me quite amazing access to senior staff, Board of Directors members for an entire day of discussions. They even videotaped me!
Many of those discussions got deep into technical weeds in a most enjoyable manner. But for those wishing to jump straight to key details, AVITI is a desktop instrument priced at $289K, a bit below NextSeq 2000, which can run two flowcells entirely independently; two sequencers for the space and outlay of one. At a cost of $1680 of consumables a user can generate 800 million reads in 2x150 format in 48 hours – or about 5-7 dollars per gigabase if running around specifications. Element believes their projected yield can be very reliably reached, with many runs over 1 billion reads. Element disclosed today three beta test sites: HudsonAlpha, the Broad Institute and Stanford.
As I’ll detail farther down, Element has partnered with multiple leading library preparation companies to enable a wide array of sequencing applications and will also offer a kit to convert Illumina libraries. They’ve also started partnering with bioinformatics tools providers.
Surfaces & Polonies
Let’s start with something I don’t generally think much about: flowcell surfaces. They’re really, really passionate about “contrast to noise ” at Element, which is basically interchangeable with “signal to noise”. Whichever way you arrange the equation, it’s about maximizing signal and minimizing non-signal. And that all starts with a surface.
As they described it, the quest for the perfect surface first means making a surface with absolutely no background - perfectly passivated as they describe it. So many iterations were tried until they had what seemed like the Vantablack of flowcell surfaces.
But such perfection is a bit of a paradox: just as a non-stick coating must adhere to a pan, the monotonous passivation perfection of the surface must be specifically modified to enable signal to occur. So further rounds of iteration, trying to provide attachment sites while minimally degrading perfect nothingness.
I asked whether the flowcells is patterned, and it isn’t. And furthermore any sort of patterning such as wells brings in even more inhomogeneity, more risk of enabling noise to latch on. Edges and corners are all special and different - and so at risk of generating background signal.
Once you have attachment sites one must prepare and attach polonies there So a next challenge was forming polonies in a reliably orderly fashion; it sounds as if Element could fill a modern art gallery with images of polony growth gone awry, but after many iterations a workable polony generation protocol emerged
Library Molecules
Library molecules for AVITI are circles and amplified by a rolling circle mechanism. Figuring out a paired end chemistry came next, and apparently gave the team great worries that they couldn’t get a robust system to work – but eventually they found one. Element and external collaborators have demonstrated nearly no index hopping and very low polony duplication rates, both sometimes frustrating on Illumina platforms particularly on newer instruments using Exclusion Amplification.
I asked about the insert size range that Element’s polony technology can support. Element has focused on typical short read library insert sizes of a few hundred bases. They didn’t believe that very short inserts (such as for micro-RNA or something like Swab-Seq) would be a problem but haven’t explored it. On the long end they haven’t looked either.
Circular library molecules guarantees not being able to just squirt Illumina libraries on, but Element will offer a conversion kit, albeit with a roughly 2:1 exchange rate unfavorable to Element – if your prep previously made exactly one run’s worth of library you’ll now need twice as much starting material. But most users will probably take advantage of native kits. On my visit day Element announced partnerships with QIAGEN, Agilent, NEB, Roche(Kapa) and Watchmaker for library kits, and in the following days they added Dovetail Genomics and 10X Genomics. Plus JumpCode Genomic’s ribosomal RNA and other abundant RNA depletion reagents. So a very wide array of sequencing applications will be natively supported on AVITI from the beginning.
Element also announced recently acquisition of Loop Genomics, which has a synthetic read technology that they sell in two forms. One kit (Solo) is intended to replace Sanger sequencing of individual clones and the other works on pools of sequences such as 16S amplicon pools. Interestingly, Element has no plans to discontinue Loop kits for existing platforms. The Loop acquisition has also meant less drive to explore longer reads or longer inserts; for these Loop is seen as the solution
Sequencing Chemistry
Once you have a library sprouted as polonies on the flowcell, how does the chemistry work? It’s an interesting twist on reversible terminator chemistry in which each position is probed twice, once with labels and the polymerase unable to extend and once with unlabeled reversible terminators to march the polony one base forwards. It also leads to an interesting philosophical question as to whether this is sequencing-by-synthesis. On one hand single base extension is an integral part of the sequencing cycle; on the other the base extension step is not what generates the signal used to resolve the sequence. In my own taxonomy, it is definitely cyclic chemistry with optical detection running on clonal molecules sitting on a surface – and I suppose I would vote to create binding as yet another mode since that is the detection step.
The beauty of this polymerase two step is in the labeled nucleotides. These are not loose nucleotides but rather multiple nucleotides are connected by linkers to a core which in turn is linked to the actual labels. What’s so clever about this is that each nucleotide interacts extremely weakly with polymerase, since it can’t be covalently linked to the precursor chain. So only if many such weak binding events happen will signal occur. Hence the term “sequencing by avidity.
These spider-shaped molecules, which the Element team refers to in conversation as “Avidites” offer many grounds for optimization – linker lengths, number of labels on the Avidite core, etc. But from a chemical standpoint they’re wonderfully modular, so it’s straightforward to experiment with one section without requiring any synthesis changes on other regions – the chemical equivalent of encapsulation in software engineering. Plus the fluor isn’t up close to the DNA, which means it is less likely to generate any sort of photodamage.
Since one is relying on Avidites aggregating many small interactions into a large net interaction, the concentration of Avidites need not be very high – nanomolar versus micromolar levels more typical for sequencing reagents. That means less expensive reagent being used in each cycle - comment was made that in typical sequencing reactions the color of reagent going in is same as that of the waste because only an infinitesimal amount of reagent is actually incorporated into growing strands. Low concentrations also means less likelihood of reagent sticking non-specifically to the flowcell.
One consequence of the avidity approach is that loss of phase results in only a very weak and slow growing background. Particularly in early cycles, within each polony dephased molecules will be few and on average widely separated. Hence wrong Avidite arms won’t make sufficient interactions to loiter and be detected.
At this time Element is offering only 2x150 kits, but with Q40 early in the read gliding down to Q30 at 150 bases. I asked if they could go longer: CEO Molly He replied that they think the core market is centered on 2x150 but they will monitor demand. Molly thinks Element is only at the beginning of exploring the new Avidity chemistry with many avenues to explore to obtain significant further optimization. With the existing quality score pattern, I do wonder if there are some select applications - such as sequencing long simple repeats for repeat expansion disorder research - that could leverage much longer reads using Element’s chemistry.
A single chemistry kit means a streamlined inventory for Element and for customers. It is indeed nice to be able to save money by ordering less expensive kits for applications that require less data, but often those savings evaporate when you find yourself with a glut of about to expire kits that don’t match your immediate applications. Actual read lengths can be adjusted in software and there will also be an option to scan only portions of the two channel flowcell to shorten total run time. CEO Molly He also believes that overall runtime can be shortened; it is set conservatively for a smooth launch but further optimization might shave time off each cycle.
Remarkably this all came together quickly, about 4 years from first acquiring lab space to an externally validated DNA sequencer.This included some nice breaks such as writing specifications for a camera and then inventing a much better one with a wider field of view and with less distortion around the edges than had been originally anticipated, enabling more polonies to be imaged with each exposure. Element also developed a new primary analysis algorithm for highly accurate base-calling of high density polonies.
Element is keeping most consumable manufacturing in house. Growth has led to three sites in the La Jolla area and one research station in the Bay Area, but the La Jolla sites will soon be consolidated to a single 80 thousand square foot building nearly completion. The new building will enable manufacturing reagents in much larger batches. And while the instrument has been launched as Research Use Only, manufacturing data systems are being built with GxP concepts in mind for the future, as well as the value of rich data for troubleshooting manufacturing issues.
As with essentially every sequencing technology company, and reflecting CEO Molly He’s background in the field, Element has a large investment in protein engineering. AVITI sports three different polymerases, each optimized for its role in the process. So one polymerase for polony formation, one polymerase for detection-by-avidity and one for extending with reversible terminator.
Data
On the data side, AVITI emits industry-standard FASTQ. Demultiplexing of barcodes is performed off instrument. Element wants to enable users to interface with any desired bioinformatics platform, cloud or not. They have been announcing partnerships in this space – with Google DeepVariant and Sentieon for AVITI-tuned variant calling models, Genoox for rare disease research, and Fabric Genomics for clinical variant interpretation.
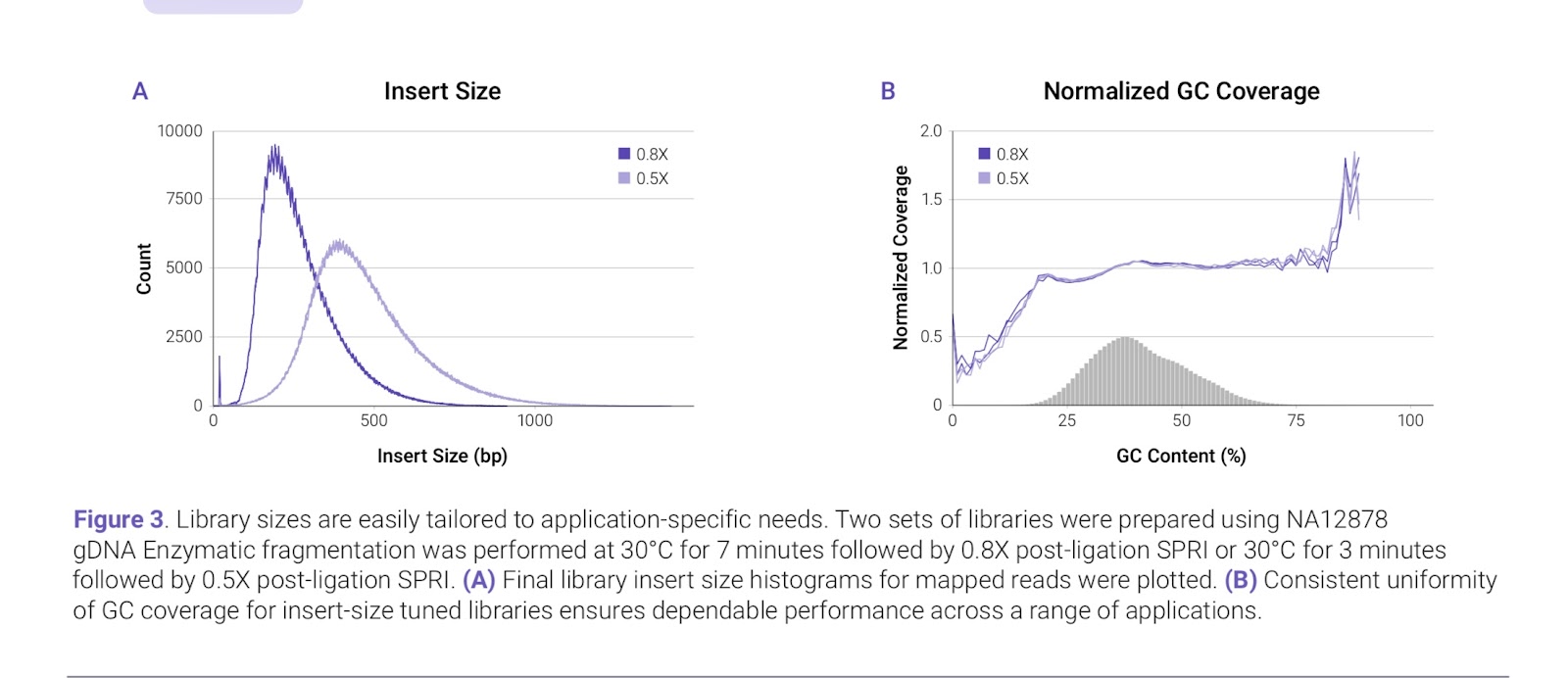
The application note from Watchmaker shows excellent uniformity across the range of GC content found in a human sample
HudsonAlpha data showed by Shawn Levy suggests that AVITI is superior to Illumina for small indel calling but slightly worse overall - but pay attention to those Y axis ticks - it’s about 0.4 percentage points different (though the failure to downsample to same depth for this slide makes the comparison dicey).

Market Strategy
In terms of competing with the entrenched market leader, Element is counting on some marketplace judo. The thinking is this: Illumina’s real focus is on NovaSeq in huge sequencing factories; that’s their main driver of revenue. AVITI will be aimed right around the overlap between the NextSeq 2000 and the bottom of NovaSeq performance (SP flowcell), but cutting under the list price of NextSeq and offering a lower (list) consumable option . And that’s not a small amount lower - based on the list pricing for NextSeq reagents currently in Albert Vilella’s wonderful NGS comparison spreadsheet, AVITI with delivers twice the output of a P2 flowcell (800M 2x150 reads vs 400M) for 47% of the price ($1680 vs. $3800) - so about 4X improvement in price vs. performance! NextSeq P3 flowcells are specified to have 50% more data than AVITI, but the price/performance ratio is around 2-fold (5-7 dollars per gigabase vs. 10.5 for P3). Of course, real values depend on field performance and whatever pricing Illumina grants an organization. Presumably Illumina could give up some margin on NextSeq to compete, but they risk disincentivizing labs upgrading to NovaSeq if the NextSeq becomes too favorable for smaller jobs - and the NovaSeq margins are too important to be touched That’s one thesis Element is working from. Element’s belief is that “decentralized sequencing will enable more scientists and clinicians to accelerate their learning without compromising cost and quality”.
Another marketing thought is lifted from the Innovator’s Dilemma: the idea is that NovaSeq really overshoots the performance needs of most labs but there is a sizable market, particularly with core facilities, for a more modestly-priced instrument that offers flexible operation but enough data to satisfy common academic sequencing experiments. NovaSeq’s temptingly low cost per base only works if you are running it in the high output modes and if those bases really are useful, and many core labs have trouble filling flow cells in a timely manner. Not only is the NovaSeq (list) about three fold the price of AVITI, but that also means the annual service contract in following years will also be about three fold higher. There’s also of course a feeling that the genomics field strongly desires some effective competition for Illumina in core markets and there is pent up demand for an alternative.
Element will be implementing transparent, uniform pricing. There will be volume discounts available, but the same ones for all – no haggling to try to get a better price only to discover you’re not as good at haggling as you think. Also good for people like me who (a) hate haggling and (b) want to be able to compare prices without worrying that reality is very different than sticker prices.
Will Element Succeed?
When I was visiting I really was taken by the focused intensity of the technical team and the enthusiasm of the leadership team – which included discussions with Board members (and industry veterans) Jim Tannanbaum and John Stuelpnagel. John, for example, catalyzed the formation of Illumina and was on its board for many years; since then he’s started or been on the board a range of genomics-focused companies in microfluidic library prep (10X Genomics), non-invasive pre-natal testing (Ariosa), immunosequencing (Sequenta) and genome editing (Inscripta). Jim’s resume overlaps with 10X, but has been more pharmaceutical focused: Jazz Pharmaceuticals, Innoviva, Theravance, and GelTex Both described in careful detail their excitement with Element’s technology and their satisfaction with how Element’s team has executed on developing that technology. On the applications side Element has snagged as a senior vice president Shawn Levy, well known for road testing new sequencing platforms and technologies during his tenure at HudsonAlpha Institute. So they have a huge base of talent and expertise to drive the launch into the sequencing market.
Will this all work out for Element? As I discussed earlier in the year, there are multiple companies jumping into the short read market over the next year or so. Singular plans to launch in early summer, BGI should be free of patent constraints by late summer and PacBio plans to launch Omniome technology sometime next year. Oxford Nanopore wants to play in this space too, though to say their reads are different in quality than Element’s is a bit of an understatement. Illumina promises higher qualities with Chemistry X, though without any details as to when this chemistry will launch and complete uncertainty whether it will be part of a new fleet of instruments. And perhaps there are other new entrants flying under the radar.
Given all that, Element will must execute nearly flawlessly in locking down some customers, getting instruments and consumables to customers, dealing efficiently and in a forthright manner with the inevitable hiccups as instruments enter the field and generating increasing buzz at meetings such as the different incarnations of AGBT. Independent confirmation of their read quality claims on a variety of different inputs will be something to watch for, particularly on challenging targets such as extremes of %GC, long homopolymers, long VNTR repeats (such as in triplet expansion disorders), and strong hairpin-prone structures.
Stay tuned. Watching Element execute promises to be interesting.
[20220315 06:16 fixed homogeneity --> inhomogeneity ]
