I'll spend most of my time talking about the NextSeq 2000, as the NextSeq 1000 seems to be a downgraded sibling with a slightly lower price -- $210K for the 1000 and $335K for 2000. Both will have the FPGA-based DRAGEN acceleration hardware on-board for faster downstream analysis -- claimed to be only 2 hours (presumably for demultiplexing and mapping a human genome). But not only will the 1000 show up later -- scheduled for late this year whereas Illumina is planning to ship 2000s and P2 flowcells this quarter -- but the 1000 won't be able to handle the larger P3 flowcell which is scheduled for later this year.
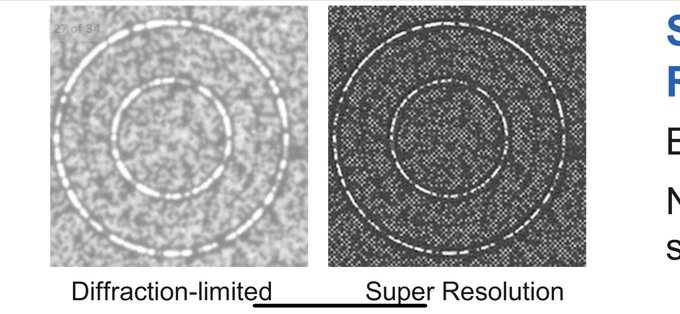
A key innovation on this instrument is the use of super resolution microscopy to image below the diffraction limit. This allows very high densities on patterned flowcells, as illustrated in an image from Illumina's slide deck; the left image is typical optical microscopy and the right image uses super resolution techniques to resolve the spots. One thing I'm curious about is just how many DNA molecules are in each of these tiny clusters? Might Illumina thinking again of going for the original single molecule concept that launched Solexa?
The super resolution approach also uses fluors which both excite and emit with relatively short, blue and green, wavelengths (dashed: excitation; blue: emission) [image from Illumina's presentation -- blame them not me for the missing scales on the axes!)
The industrial design of the new NextSeqs seems to have generally generated favorable response, though as is the norm for big brothers, from that front some snark.
I think my new sibling has inbuilt #PhillipsHue because you need to have the right ambiance to sequence. #SBS #PastelColours #innovation pic.twitter.com/IwDFl3s0tD— NovaSeq6000 (@seq6000) January 13, 2020
In terms of output (Full specs ), it's useful to use 2x150 as the comparator since Illumina supports that on all flowcells. The P2 will over similar yield (approximately same number of clusters) to the NextSeq 550 Hi Output kit for a similar run time. How the pricing will compare hasn't been released. The P3 will deliver a cluster count intermediate between NovaSeq's SP and S1 flowcells, though with a much longer run time than either.
That seems to expose the catch for super resolution is apparently slower cycles and therefore longer runtimes. The P3 chip will have 1.00B clusters vs. the 1.45B clusters on a NovaSeq S1 flowcell, yet for 2x150 the projected runtimes are 48 hours for NextSeq 2000 P3 vs. 25 hours on S1 -- clearly something is slowing down the scanning. By my estimate (estimating fixed time for clustering and paired end flipping at 7 hours), it looks like 8.2 minutes per NextSeq 2000 P3 vs. 3.6 minutes on NovaSeq S1. Or look at 2x50 mode: the P3 chip will deliver less than a third the data of an S2 chip but take three hours longer.
In terms of markets, CEO Francis de Souza described the new instruments as the "natural heir to NextSeq" and particularly noted "in oncology a drift to larger panels". Sequencing human trios was also cited. de Souza repeatedly described the new instruments as ideal for clinical situations Illumina is seeing, so while there isn't a DX version announced yet (though a NovaSeqDX was announced), one can bank on one showing up, maybe in next years JPM presentation. Launching these instruments is a bit of a shift from Illumina's previously professed strategy of convincing NextSeq owners with high utilization to upgrade to NovaSeq. Perhaps they have market research that some of these companies prefer desktops to the large floor model NovaSeqs, or they just felt that having a lower capital instrument would sell. Or this was a good place to trial the new super resolution technology.
That seems to expose the catch for super resolution is apparently slower cycles and therefore longer runtimes. The P3 chip will have 1.00B clusters vs. the 1.45B clusters on a NovaSeq S1 flowcell, yet for 2x150 the projected runtimes are 48 hours for NextSeq 2000 P3 vs. 25 hours on S1 -- clearly something is slowing down the scanning. By my estimate (estimating fixed time for clustering and paired end flipping at 7 hours), it looks like 8.2 minutes per NextSeq 2000 P3 vs. 3.6 minutes on NovaSeq S1. Or look at 2x50 mode: the P3 chip will deliver less than a third the data of an S2 chip but take three hours longer.
In terms of markets, CEO Francis de Souza described the new instruments as the "natural heir to NextSeq" and particularly noted "in oncology a drift to larger panels". Sequencing human trios was also cited. de Souza repeatedly described the new instruments as ideal for clinical situations Illumina is seeing, so while there isn't a DX version announced yet (though a NovaSeqDX was announced), one can bank on one showing up, maybe in next years JPM presentation. Launching these instruments is a bit of a shift from Illumina's previously professed strategy of convincing NextSeq owners with high utilization to upgrade to NovaSeq. Perhaps they have market research that some of these companies prefer desktops to the large floor model NovaSeqs, or they just felt that having a lower capital instrument would sell. Or this was a good place to trial the new super resolution technology.
Which is the basis for a prediction I'm making today: I believe this super resolution technology will expand, particularly at the high end. So I expect that next year's JPM presentation will include an upgraded NovaSeq design. These would require new optics, but might have 4X the yield for each flowcell and perhaps have a legitimate claim at a $100 clinical depth human genome. But given my math above, the cycle time might make a huge jump to the already circa 7.4 minute cycle time for an S4. If my math is right, we might be seeing 2x150 on an S16 flowcell (following Illumina's naming pattern; each step up is a doubling of number of clusters) might run longer than three days.
If you were just looking at output, there's still a bit of a jump for 2x150 between 7 gigabases on the MiniSeq Hi Output kit and 36 gigabases on the NextSeq 550. So perhaps next year will bring a super resolution MidiSeq spanning that range? That's three possible predictions for a year from now -- I'm really getting ahead of myself!
If you were just looking at output, there's still a bit of a jump for 2x150 between 7 gigabases on the MiniSeq Hi Output kit and 36 gigabases on the NextSeq 550. So perhaps next year will bring a super resolution MidiSeq spanning that range? That's three possible predictions for a year from now -- I'm really getting ahead of myself!

1 comment:
This is, in part, a testing ground for the new chemistry/SRM approach - just as the original NextSeq was for the two-colour chemistry. It took a little while for that chemistry to be optimised. Anyone who experienced v1 NextSeq data knows it wasn't good. I assume they'll work out the kinks and then incorporate it into future sequences too.
It's also telling that although they weren't sure,t kept the NextSeq branding. NextSeq output fits well with most clinical labs. 20-120Gb is perfect for small/medium panels of scores of patients at reasonable depth. I know of several clinical labs with at least a couple of NextSeqs for this purpose. I guess they still want to focus on these labs, driving down the cost. DRAGEN also feeds in here, making analysis faster and more streamlined.
What was slightly disappointing was the small dynamic range of flow-cells available. One of the great things about NovaSeq is the highest output is more than an order of magnitude greater then the lowest output. There's still a gap at the low end between MiSeq and NextSeq which could have been filled with a SP flow cell with, say, 50M clusters.
Post a Comment