Blog team member intently watching @Clive_G_Brown webcast - now must confer & write-up impressions pic.twitter.com/jPGpw1w0lg
— Keith Robison (@OmicsOmicsBlog) March 14, 2017
GridION X5
GridION X5 is intended to serve labs that see the MinION as too small but the PromethION as overkill (more on the big beast later in this post). Sporting five flowcells and hefty on-board compute, it is intended to replace the snarl of cables and machines seen in larger MinION labs. GridION is planned to launch on May 15th, just after the London Calling confab. Clive swears that manufacturing is well underway and that this design can be produced in bulk.
GridION X5 and PromethION licenses will both permit use in a service capacity; it will now be legitimate to profit from sequencing other people's samples. Clive didn't elaborate on the logic of blocking MinION users from this, saying only he and Spike (Wilcocks, the ONT commercial point person) understood the logic. I'll agree I don't, but presumably it is more of an internal commercial decision than anything anyone on the outside would cook up. In any case, Oxford will also be launching a certified service provider program.
Clive didn't dwell on operational details, as it was late in the presentation slot by this time (I've reported this presentation according to my own peculiar order, not the temporal one). Presumably the five flowcells are fully independent.
.
GridION X5 is a huge step up in cost over the MinION. There are two pricing schemes, one treating the instrument as a capital item and pricing flowcells more cheaply and another "reagent rental" pricing scheme. The upfront option costs $125K but then you can buy up to 450 flowcells over the next 18 months for $299 each. The reagent rental option opens with only a $15K training fee (included in the purchase option) upfront, but requires a commitment to 300 flowcells at $450 each.
I've plotted out the cost curves (below) for these and am scratching my head; the purchase option (blue triangles) is only better for 100 or fewer flowcells than the reagent rental (red circles). I'm wondering if someone at ONT didn't quite do their sums correctly -- or perhaps I haven't? I would have at least thought the flowcell discount would disappear with at least breakeven versus the reagent rental pricing, but you can see from the jump in the curve this isn't true.
Ideally I'd have a curve for MinION there, but I don't know the cost for the equivalent of the compute included in the instrument. It's a nice box - "latest gen CPU", custom FPGA (more on those in a bit), 64Gb RAM and 8 TB SSD, but $125K seems high. The lowest MinION flowcell cost is $500, so the slope would be steeper. If you want to run as a service to external customers, or just like a neater look than some 3D-printed MinION holder and cables to your compute, then GridION X5 might be worth it.
PromethION
Clive described PromethION as the elephant in the room, as the PEAP (PromethION Early Access Program) has been far behind schedule. This prompted me to write a very skeptical piece late last year, wondering if PromethION had been a mistaken detour. Clive promised that somewhere between 12 and 20 of the PEAP sites would receive four flowcells which will ship on April 3rd. Clive noted that PEAP users have been very patient, with perhaps just one customer requesting refund of their deposit.In addition to kits going out, PromethION will be undergoing a complete makeover. Oxford's engineers decided that cramming all of the compute inside the sequencing box just wasn't working out, so a computing unit in tower configuration will be coming out. With FPGA coprocessors, this beast will have up to 80 teraflops of power, which Clive believes will enable real time basecalling even when the machines are fully configured to 48 flowcells and the pores run at 1000 bases per second. ONT is also planning to implement their Epi2Me software in this system, so that users averse to cloud computing can run all their workflows on spare capacity within the tower unit.
After tower launches (scheduled for Q3), then PromethION will slim down, as the lower section will contain only a networking switch. This would enable connecting to the tower unit, as well as to local compute power. Around Q4, Oxford expects to enable PromethIONs to run all 48 flowcells; the slimmer case would probably launch around the same time. Eventually, early next year, the entire box will be re-engineered to accomodate all that is learned during PEAP. That future product is already designated PromethION MkI.
On the manufacturing front, Clive stated the backlog of PromethION orders should be cleared by Q3 of this year, though he lamented he had stated a year ago that this milestone was to be reached in Q3 of 2016. Flowcell manufacturing is scaling up as well, as seen in the tweeted image below
CB: PromethION early instruments continue to ship, flow cell production underway #nanoporeconf pic.twitter.com/xPQOQLRb7B— Oxford Nanopore (@nanopore) March 14, 2017
Throughput
Clive threw around a lot of throughput numbers for all the instruments, but perhaps more interesting is that Oxford is now putting a team on the discrepancy between yields obtained internally and those seen in the field, which are at least 2X and perhaps much worse (in particular, I seem to emit some sort of MinION dampening field). Oxford's belief is that low yields are largely due to users starting with contaminated, sheared or poorly quantitated DNA. So ONT will be exploring the limits of these problems to nail down their effect sizes.Longer flowcell lifes are on the horizon; Clive has tweeted out results of 60 hour runs. Long-lived pores need good libraries though.
ONT recently increased yields by improving the solution to pore jamming. A new release of MinKNOW will shorten the detction time for blocks to under 2.5 seconds; currently this is around 5 seconds. A bullet suggests that better QC tools integrated into MinKNOW are coming, an idea I suggested earlier this year. ONT's Dan Turner announced today a new command line tool called 'wub' which will apparently provide QC metrics on runs.
1D^2 Chemistry
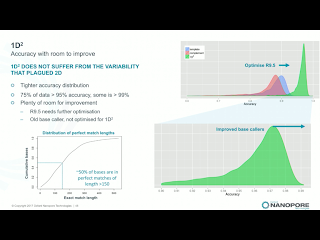
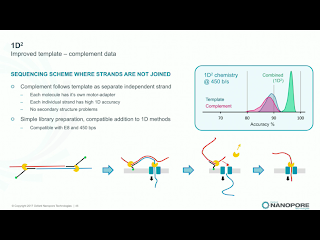
Clive announced that 2D library prep kits will be discontinued on May 5th. While he made no mention of it, the 2D discontinuation seemingly ends the point of dispute in Pacific Bioscience's intellectual property actions against ONT. Instead, the new 1D^2 chemistry will be rolled out along with a slight change to the pore, R9.5. R9.5 chemistry is promised to be fully backwards compatible with all R9.4 callers and kits. 1D^2 has not physical linkage between the two strands. Instead, as the first strand is pulled through it tends to drag the 5' end of the second strand towards the pore (illustration below). With R.9, about 1% of the strands show the 1D^2 phenomenon, in which the second strand is acquired by the pore almost immediately after releasing the first strand. I've seen a few "mirror reads" in our data, in which the software does not recognize the brief free pore state and treats both as one long read. The R9.5 pore increases the 1D^2 phenomenon to around 60% of reads, and Clive leans towards the explanation that nicks or failure to ligate an adapter are significant contributors to the 40% of reads which do not exhibit the effect.Holders of developers' licenses will have access on the 27th of this month to custom flowcells with the R9.5 chemistry and the prototype 1D^2 basecaller, with community release of 1D^2 at the London Calling meeting in early May. Longer lived flowcells based on the R9.5 chemistry will apparently ship on April 24th, a bit before 1D^2 is available. Clive suggested that kits may be available which lack the 1D^2 capability, for users with applications in which total independent sequence is more important than the higher accuracy. Side thought: since 1D^2 happens at a low level in any case, it will still be useful for the basecallers to recognize this so that the non-independence of the reads is recognized and so that the monster pseudo-hairpins I've seen are resolved into 1D^2 reads.
Basecalling
The long-hinted axe on cloud basecalling drops on March 21st. Clive noted that the cloud basecalling was very useful for developing the basecalling software, but once local basecalling was made available it was left in place as a safety net. That net is going away; Clive remarked that if you leave a safety net in place "people start using it as a hammock". Oxford will be releasing new compute specs shortly; with computers from this list it should be nearly possible to run local basecalling in sync with sequence generation. A new tool called Lamprey will allow users to effectively mimic the cloud basecalling system on their local cluster.Clive presented some error numbers based on a 20X coverage assembly of E.coli. The first slide below shows the consensus error profile for this assembly using the current basecaller and either 1D or 1D^2 chemistry. Curiously, the 1D^2 actually does worse on mismatch and insertion (!), but improves on deletions in both homopolymer and non-homopolymer cases by 2X-3X
The yet unreleased Scrappie basecaller relies on a "transducer" model. Rather than being released separately, this will apparently be rolled into production basecallers on April 20th. The data provided for Scrappie on the E.coli assembly is only for the 1D chemistry, but it improves mismatches by a factor of 4, non-homopolymer deletions by a factor of 13 (!!), homopolymer deletions by a factor of 4 and a very small (~6%) improvement on insertions.
ONT had remarked before that Pacific Biosciences had pointed out ONT's homopolymer issue in a presentation, showing a case in which a polymorphism has been implicated (well, Clive disputed this) in Alzheimers. With newer chemistry and basecallers, a heterozygous sample is correctly called from the consensus. Note also the belated appearance of Monty Python references; both prior webcasts had them in the titles!
A huge upcoming change in the basecallers is the elimination of the "event" model, and in many cases elimination of the FAST5 files altogether. The current software parse the raw electrical signal into discrete events which are stored in the FAST5. Clive has apparently wanted to ditch the events for some time, as he has long felt it was a poor model. With the newer basecallers, the option to generate FASTQ or unaligned SAM will be available, or users will be able to get FAST5 files which have just raw signal -- and are promised to be over 80% smaller. Developer releases Scrappie with the new scheme will occur during Easter.
FPGA Basecalling Accelerator
Clive has previously broached the idea of a Field Programmable Gate Array (FPGA) approach to accelerating basecalling. Working in collaboration with Intel, Oxford is continuing to work towards a PCIe FPGA card which could go into PCs, as well as a USB device to support MinION on laptops. The USB device is farther out and ONT is not yet sure if it will be inline with the MinION or occupy a second port. As shown below, basecalling speeds of 50Kbases/s are achievable with a desktop CPU hitting 0.5 teraflops, but only 10% of that power is actually used. As Clive explains it, the recurrent neural networks now being used for basecalling are challenging to parallelize, as information must feed back-and-forth through the network. With an FPGA it is possible to achieve 60% utilization of a 10 teraflop rated device and hit 7.5Mbases/s calling with the same power draw as the desktop CPU. PromethION is slated to deliver about 65Mb/s, so a set of 10 such FPGAs should be able to keep up. Clive also stressed that FPGAs can be scaled up-and-down; the solution for SmidgION and Flongle can use a smaller circuit, for MinION a bit bigger and for PromethION a really large one.
Quick Flashes and No Updates
Clive gave a number of small updates, a few things showed up only in his release schedule slide and others got a single sentence. Many other items were simply mentioned as exciting but no updates until London Calling. I will bang out most of these as 1-2 sentencesClive is planning a third go at the CliveOme, this time using megaread protocols. He is holding out hope of getting a centromere-spanning read or even an entire chromosome. ONT has an effort to extend leviathan reads to entire chromosomes (I think yeast was mentioned).
Approximately 4000 MinIONs have been shipped!
VolTRAX chemistry should ship on March 27th. VolTRAX multiplexing planned release on May 15th.
Direct RNA early access is listed as March 20th; a bit of a puzzler since there have been tweets of direct RNA results. But perhaps those were just 1st trials and this represents a reliable supply.
Essentially no news until London Calling: cDNA (though Clive said it is spectacular), sequencing from 20 pg input via whole genome amplification, barcoding kits (I'm hoping Oxford goes beyond 12 barcodes; that's only a good start!), Zumbador, Flongle, SmidgION and Metrichor/Epi2Me.
Okay, that's all. Hoping my accuracy is better than my sometimes garbled reporting on leviathan reads! Can't wait for London Calling!
[Jared Simpson found I wrote $399 instead of $299 for the flowcell price in the upfront GridION X5 pricing scheme; my graph generator had the correct number but now the text is fixed 2017-03-14 23:41]
[Vinzenze Lange pointed out the original cost curve plot was off; I botched the change in slope after the flowcell discount runs out 2017-03-15 06:32]
[Vinzenze Lange pointed out my text reversed "red" and "blue" -fixed 2017-03-15 10:47]
Should have put this in originally - Python code for the plot
import matplotlib
matplotlib.use('Agg')
import matplotlib.pyplot as plt
own=125000;
rentCost = []
ownCost = []
fcN = range(0,600,10)
rentBase=14999;
ownBase=125000
for fc in fcN:
if fc<=450:
ownCost.append(ownBase+299*fc)
else:
ownCost.append(ownBase+299*450+(fc-450)*475)
if fc>300:
rentCost.append(rentBase+475*fc)
else:
rentCost.append(rentBase+475*300)
rentPlot=plt.plot(fcN,rentCost,'ro',label="Rent")
ownPlot=plt.plot(fcN,ownCost,'b^',label="Own")
plt.ylabel("Total $")
plt.xlabel("Flowcells")
plt.ylim(0,350000)
#plt.legend(handles=[rentPlot,ownPlot])
of = open("/home/krobison/www/plots/test.png","w")
plt.savefig(of,format="png")









32 comments:
Great post!
Purchase option is blue (not red) triangles in your cost comparison. (and likewise red circles for the rent option)
I do not understand the quite big jump in your graph for the purchase option after the first 450 flowcells. However, that does not change the general message that the purchase option seems only to be sensible for grants that treat consumables differently from purchases.
Vinzenz:
There should be a change in slope there as the flowcell cost goes up, but you are correct that I made a mistake -- corrected plot posting in a moment
The CapEx option costs 125K but you are exempt of paying the 15K annual service on the 1st year
that makes the like start a bit lower ... doesn't it?
Duarte:
Code looks right - you are correct that the support/training fee is included in the $125K purchase option
Keith ... I beleive your price for the flowcells is incorrect in opex is it 475 not 450.
Also where did you see that the discounted 299 flowcell price was only for the first 450 cells?
it does not say that on their website
https://nanoporetech.com/products/gridion
you Are correct however. If there is a limit for the discounted flowcells then the capex mode makes no financial sense.
also... the opex mode requires the customer to buy at least 300 flowcells. The CapEx mode does not have a lower minumum flowcell order... so in truth if you want to run less than 300 flowcells you would need to go for CapEx so the charts do not start at the same point
Assuming no Capex limit on the discounted flowcells the breakeven point is at about 600 flowcells in a 3 year timeframe
https://docs.google.com/spreadsheets/d/1YpKZp9WP8YmNaBfYtpklulQJCdLH6XcmJ_XzR9zMdls/pubchart?oid=522361220&format=interactive
Duarte:
Clive's slide had the 450 limit on it. You are correct that the website doesn't mention this -- an important point & I'll ask ONT for clarification
Albert Vilella has looked at my chart and he suggested that the CapEx might include for free the first 300 flowcells
so the price would remain fixed up to 300 flowcells then the flowcells would cost 299 up to the 750 flowcell purchased (300+450) and then revert to 475 for the remaining ones
This would make the CapEx much more atractive
https://docs.google.com/spreadsheets/d/1YpKZp9WP8YmNaBfYtpklulQJCdLH6XcmJ_XzR9zMdls/pubchart?oid=522361220&format=interactive
Where does the information about MinION/GridION flowcells not being compatible come from?
Lars,
That's my inference; I could be wrong (I should check with ONT). Clive's slide says "Based on on current MInION flowcell design", which I took to mean they are not MinION flowcells. But perhaps someone just chose odd wording on the slide
I read that as 'using existing MinION flow cell design'. The products page explicitly says "Use up to five MinION Flow Cells at a time" https://nanoporetech.com/products
Lars,
Thanks -- will update! Wish they had been explicit like that on the slides.
So 5 minions at $1000 or less each equals one X5 at $125k. Sure there is some compute cost in the GI but $120k? Surprised that there has not been more chatter about this. When you compare apples to apples (ie Q30 data) a GI costs slightly more than a Miseq and produces about the same output in 3x the time (assuming an optimistic 20G per flowcell in 60 hrs and 20 fold coverage required to reach Q30). Hmm...
Will you be writing a post about the "sequencing with nanopore using n-mers" patent that pacbio was awarded? I'm really curious to read your analysis on this one! It's pretty crazy that a company who doesn't even use nanopores managed to jump on the patent before anyone else.
No, it is very common actually. Just google "patent troll".
Surely it's more throughput than miseq. Thought miseq was 15-20G in 24 hrs ? The recent MinION runs were 20G in 48 each. They're claiming to double that again soon. So surely 5 of them is ~= 2 miseqs ? This also ignores sample prep time, quick on nanopore. What's in a miseq ? A camera and a pump and a PC and glass flowcells.
PacBio's "sequencing with nanopore using n-mers" is really strange. They are claiming the same stuff that Clive showed in the original minIon unveiling slides and this was filed 3 years after that.
The patent it extends from 2010 looks like a basic, general description of nanopore sequencing that was already covered by patents from the 90s. How were they awarded any of these?! I'd love to hear from someone better informed.
"PacBio's "sequencing with nanopore using n-mers" is really strange. They are claiming the same stuff that Clive showed in the original minIon unveiling slides and this was filed 3 years after that.
The patent it extends from 2010 looks like a basic, general description of nanopore sequencing that was already covered by patents from the 90s. How were they awarded any of these?! I'd love to hear from someone better informed."
The USPTO is very sloppy. It allows things through that other patent authorities don't, then they are settled in expensive lawsuits. Almost like a tax on doing business in the US. For example, EU PTO an Japan have stronger filters on patent allowance upfront. The patents in question have not been enabled by Pacbio and are concepts, the key claims were appended following ONTs AGBT 2012 talk. People can draw their own conclusions about the fairness of that. My reading is that ONT don't practice any of the patents Pacbio is suing them on anyway. It is likely therefore that either Pacbio don't understand the technology, or are resorting to litigation as a fig leaf, knowing full well they are rolling dice.
I saw Alexander Wittenberg's tweet about the Sequel only managing 5-6 GB and then Nick Loman's tweet, almost the next day, of a 5 GB Minion run that had 800kb reads in it. That's hard for PacBio to compete with on a science level. Couple that with the fact that anyone with $1000 can do it, not just facilities that can stump up $350k and the required space for a Sequel, then we can all see the writings on the wall. I know PacBio promise there will be 32x more throughput by the end of 2018 but that is in 20 months time, look at what Nanopore have done in the last 20months, how far ahead they will be by then?
To take an action that could be said to have a whiff of patent trolling indicates to me time is short and they are desperate. Will they be here in 20 months to deliver 32x?
That makes the logic of a district court action seem odd as they take years to conclude, by which time PacBio will likely be long gone.
My guess is they know times is short and their only option to save themselves is an acquisition. They can't appeal to Roche or Illumina with their product (remember Roche dumped them) but they can try to make out that buying them offers the buyer a Nanopore torpedo.
That is my guess anyway.
To the poster above me:
Your comparison isn't apples to apples. With current ONT 9.4 and basecalling methods it takes roughly 30x coverage to reach QV20. It only takes 5x coverage of pacbio sequencing to reach QV20. And if your using Pacbio CCS you hit >QV20 with just 2 passes. So if you're looking to assemble a 1GB genome with QV20 you would need 30GB of nanopore data or 5GB of pacbio data. Just looking at raw output without looking at error rates is useless when pricing the cost of a genome. This is why pacbio still dominated the long read market and (currently) ONT is not more cost effective. Both companies claim major upgrades in the next year so we will have to wait and see how it plays out.
On the other hand I agree with you about the patent trolling. I really wonder what's going on in the mind of pacbio execs. It seems like the past few months have been filled with poor decisions (including losing the roche contract). Maybe they are using these patents to increase the company value for a sale.
"Your comparison isn't apples to apples. With current ONT 9.4 and basecalling methods it takes roughly 30x coverage to reach QV20. It only takes 5x coverage of pacbio sequencing to reach QV20. And if your using Pacbio CCS you hit >QV20 with just 2 passes. So if you're looking to assemble a 1GB genome with QV20 you would need 30GB of nanopore data or 5GB of pacbio data."
You make a very fair point, although I thought the latest data releases showed 15x was required for QV20 with MinIon (I will try to dig out where i saw that - I think Jared Simpson but please say if anyone knows to save me effort haha)? With that in mind and if you believe company specs (again haha); PacBio claim sequel is 9 GB and Nanopore 20 GB. Therefore I stand by my point that on a science level the Sequel just can't compete (again there is also cost and practical considerations that work against Sequel).
A side point but why do PacBio make Lego toys, surely it just illustrates what a behemoth the Sequel is!? Their PR department need sacking for that as they look stupid when their competitor is actually tiny (gimmicks irritate me but badly thought out gimmicks... urgh)?!
I don't believe company specs. A fair comparison is Pacbio customers get ~5-8GB/run while ONT customers get 1-10GB/run. So far no customer of ONT are getting 20GB. That's an upgrade planned for the future. But Pacbio also said they are increasing the throughput to 20GB/run this year. Until customers actually start getting those values there's no point of claiming that's what the technology can product. We need to compare what customers are getting now.
I believe the 15x QV20 ONT data was provided by ONT about their updated basecaller. I don't think any customer has produced that. Even if it's true that still means you need 3x more ONT data to produce a similar quality genome as Pacbio.
I think the lego toys ended up being a big hit at AGBT. But it's not like that will sell them more instruments. Management seems to handle major decisions pretty poorly.
Ah ok, thanks for clarifying.
I guess it will be interesting to see how fast each progress. Sequel seems like the only major upgrade PacBio has done in 6yrs so not sure I'm convinced by their claim of a further 32x in 20 months.
Contrast that with the 20 month lifespan of MinION where they have managed to get higher throughput than a Sequel on something that is actually Lego size already (although as you contest throughput isn't everything). It has also done it with reads of nearly 1 Mb in length.
I guess I just hope it is the end of fluorescence, based on a love-hate relationship of growing up with it, haha.
I'm actually confident they can reach 32x by the end of next year. Pacbio increased the throughput of the RS by about 100x over 4 years and I'm sure they can continue that on the Sequel. Their technical team is top notch. It's the management team that seems to be lagging. It's a shame because what they have accomplished technically is amazing. Too bad their management team has been making poor decisions (with this new patent lawsuit being the most recent). I can't believe Mike H. actually thinks they could win this case. What a waste of money for everyone. Maybe the board will get rid of him. He lost the Roche deal and now he's spending resources on patent trolling lawsuits. What a joke.
@Ben, unfortunately it appears you are making your case to someone who appears to be paid to make these comments - At least I would be skeptical of someone who scripts comments with lines such as 'You make a fair point and 'on a science level...'
In spite of Keith's best intentions, the comments section can be a bit of an echo chamber and if you are arguing with an active paid commentator who is not actively involved in the tech and whose sole purpose is to make sure ONT always looks good, there is very little actually learning from the discussions in comments.
One way to improve this is to make commenting truly non-anonymous but that comes with its own pitfalls.
Not a paid commentator at all, nor am I a consultant in any way. I admit that as a Brit I am heavily biased, however.
I am also bored of the same old, same old SBS though. Nanopore is a fundamentally different approach and that is refreshing. I also think that is the reason PacBio can't and doesn't compete on a science level.
Hi Ben,
Are there any paper or preprint that shows how much coverage is needed for Sequel and R9.4 to achieve QV20? Thanks a lot in advance.
Here is the coverage data on Sequel: (~5x coverage needed for QV20)
http://www.pacb.com/smrt-science/smrt-sequencing/accuracy/
If you use their CCS method you only need 2x coverage:
https://www.omicsonline.org/articles-images/data-mining-genomics-Read-quality-value-4-136-g003.png
The recent human sequencing effort with minion showed ~30x coverage to get 99% (Qv20) accuracy:
https://genomeinformatics.github.io/NA12878-nanopore-assembly/
Hope that helps.
Isn't CCS essentially just coverage though, but in a form with a vastly lower read length?
Thanks Ben for your quick reply.
I think PacBio still has an edge when very high base accuracy is required. Also, PacBio's methylation stuff is at production level whereas ONT remains research grade.
I would not say is doomed on a science level as long as they can still crave out a big enough niche to keep them survive commercially.
I think ONT's technology depends heavily on the protein they use to make the pores. In this R9 generation, they managed to improve accuracy and throughput significantly such that PacBio's niche is further reduced.
The problem of ONT is they need to find new protein that makes better pores. This is more a hit-and-miss process. On the other hand, PacBio's progress can be more steady as it depends more on advances in optics and compute.
ONT claims they have an new R10 protein. Let's wait and see what it brings. I hope PacBio can also come up with something even better soon. I think most scientists would rather PacBio stay long to serve as an alternative and a competitor to ONT.
More details on the GridION. CapEx. You only get 18month discount. OpEx you have to use 300 flow cells in 12 months that is about ~60% of capacity assuming 5 day work week and 50 weeks.
It hard to look at the GridION as anything but a massive service provider fee that doesn't go away. What happens in year 2? do I have to buy another 300?
With MinION I could buy 5 for 5k (assuming ONT would sell them to us) and a 48 pack to. That would get us to a $500 discount level for a total of 29k vs 157k for OpEx (300 X 475)+ 14999 service contract))
So, how are their sales in comparison to PacBio? Are they catching up?
Post a Comment