Chemistry & Basecalling
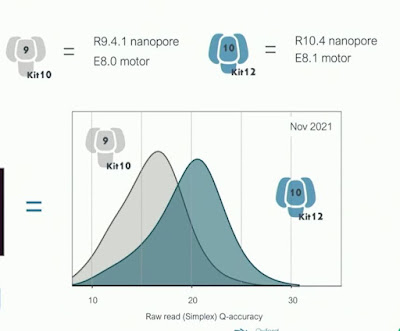
One of those exits is enabled by a combination of a new chemistry and a (wise IMHO) decision by ONT to simplify their offerings. Clive announced a new chemistry, kit 14, which will soon be available and which combines an R10.4.1 revision of the R10.4 pore currently available with a new motor E8.2. Kit 14 will offer the high accuracy of the Q20+ chemistry, enable duplex sequencing and base modification calling. A surprise that Clive dropped is that this is the beginning of the end for the venerable R9 pore - ONT will offer only a single pore chemistry going forward. Phase-out will be gradual with R9 chemistries available through the end of next year to enable large projects to finish up, but customers are strongly encouraged to switch ASAP. That is if they can -- there are still in the field early access PromethIONs which cannot use R10 flowcells and ONT hasn't prioritized arranging upgrades of these museum pieces.
The new motor inherently enables somewhat faster speeds and a several favorable properties for high accuracy basecalling: tighter speed distribution, better resolved signal levels, and fewer misstep , E8.2 arose from an extensive search for mutants that performed well with existing basecalling algorithms, which were then re-turned on the promising mutants. To support both high accuracy and high yield use cases, instrument software will enable controlling the flowcell temperature - perhaps even during a run. Higher temperature, faster translocation and more data at lower quality; lower temperature, less data but higher quality. - though Clive stated that the "tradeoffs are quite multidimensional and complicated" for temperature control. I had wanted to play with this dial back when the MinION Access Program (MAP) was launched in 2014 (between weak flowcell supply and difficulty getting data and it wasn't supported, that concept died quickly then) - glad to see it become reality.
Get out some old newspaper: it's time to wrap up Guppy. A new basecalling tool called Dorado will replace it and will make it's initial splash at London Calling. Dorado will be open source, optimized for GPUs and PyTorch. With Dorado and A100 towers it is expected that the P48 PromethION will be able to keep up with basecalling even in High Accuracy Mode (HAC) with Clive dangling the possibility that Super Accuracy Mode (SUP) might also be able to soon keep up. Dorado will also support Apple Silicon.
Clive also mentioned the in-browser version of Bonito which via the magic of WebAssembly enables accessing the latest-and-greatest version of the basecaller on your local machine - no data is transferred over the wire. I should try this again; my first attempts never seemed to ever do anything.
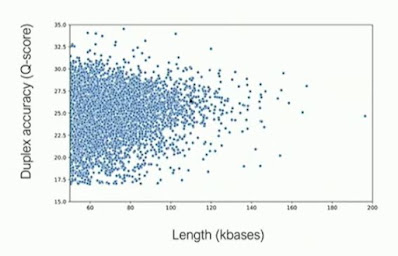
Clive gave a few updates on duplex sequencing - reading both strands of a molecule and generating a joint basecall. He mentioned that "follow-on", the second strand just naturally jumping in after the first finishes, occurs at about a 5% rate without encouragement. With "follow on" adapters, whose architecture he did not describe but it would seem this is a non-covalent interaction, can raise this above 50% in ONT's hands. The key advantage ONT wants to tout is no length dependence (vs. the current limit of PacBio's HiFi scheme at around 25 kilobases), and his plot showed duplexes and their quality for read lengths in excess of 100 kilobases - albeit with an accuracy distribution significantly downshifted from what would be seen in HiFi data (not that PacBio has their accuracies perfectly calibrated either) - HiFi data has large fractions of reads simply above the top of the Y-axis in the below plot. But I definitely have projects where I would salivate over 200 kilobase Q25 reads, so duplex sequencing looks very exciting. Some devil in the details: which library prep workflows will be initially supported for duplex sequencing?
An interesting reminder from Clive that current Q-scores from the basecallers aren't particularly useful. ONT is working on recalibrating the Q-scores so that they will be useful guides to the accuracy of the data and particularly for identifying low quality islands within otherwise high quality sequences
On the adaptive sampling front, Clive noted a number of performance improvements that mean on GridION the keep/discard call can be made using 200 basepairs. Adaptive sampling is now working in beta on PromethION, though with a longer response time that ONT believes can be improved to the 200 basepair speed with an imminent firmware update.
Clive also went quickly over "Outie" chemistry, ONT's scheme for getting even higher accuracy by flossing the DNA multiple times. No timing was given on when anyone outside ONT could play with this. He describes this as for "needle in a haystack" applications such as rare variant finding.
The original theme of the talk was saying Me, as in Me-thyl. This led to a moment of unintended humor - Clive accidentally pronounced the modification in the American way (rhymes with Ms. Merman's first name ) rather than the proper British version (rhymes with "wee isle"). Plus the Knights Who Say "Meh!" isn't a way to get people excited about anything. Anyways, once Clive corrected himself he made the bold claim that for 5-methyl-C identification bisulfite isn't a gold but bronze standard -- and ONT is now better than bisulfite. I doubt anyone would be disheartened to eliminate a protocol with a mutagen in it, but it will probably take some testing by an outside group such as ABRF before ONT can truly claim gold. There was also some interesting discussion on Twitter on whether PacBio's 5-mC calling or ONT's is more accurate. Again, some neutral referees are needed to sort this out.
An interesting peek under the hood was given of how ONT is training their modification models . ONT synthesizes libraries consisting of a block of known sequence, 30 random bases, the modified base, 30 more random bases and then another known stretch - and then ligates this to carrier DNA and sequences it. It's a bold approach to generate training data that shows their confidence both in the baseline basecaller models as well as their training methods. After all, oligo synthesis has its own spectrum of errors dominated by deletions so the actual position of the modified base will slip a bit.
Short Fragment Mode (SFM) has now been released - though with Clive lamenting the slide section was still labeled "Short Read Applications". This is a set of software improvements to properly support reading short fragments. ONT hopes to use this to wean users away from short reads in counting applications. Whether many buy into this remains to be seen.
Clive again submitted to being "the research substrate" and provided blood for demonstrating SFM on cell-free DNA, showing the characteristic pattern of fragment length peaks corresponding to nucleosomes. And since this was native cfDNA, methylation calling is on the table.
Hardware
On the hardware side, the key releases are the two-flowcell miniature "P2" versions of the PromethION, one at $60K which has onboard compute and a compute-less version for $10.5K that is expected to be purchased as an expansion by GridION users since the GridION can loan out its GPU resources to its new sidekick. P2 will have adaptive sampling from the start.
P2's big siblings are getting a compute upgrade; from May onwards the $225K P24 and $310K P48 will ship with A100 GPU towers. This will make uniform the compute available on the two instruments rather than P24 having half the power, and in addition to the newer edition GPU will have 512 gigabytes of RAM in place of 384 gigabytes before (did I ever mention my first two computers started with 1K of RAM?)
Finally on this front, four lane flowcells for PromethION are returning to the lineup.
Clive reiterated his prior enthusiasm for the MinION Mk1D, a custom keyboard-bearing case for an iPad Pro that has a slot for a flowcell plus a region that may in the future host a VolTRAX or similar device. And it is truly "just" a case; the iPad Pro has sufficient horsepower to handle the basecalling and other computational needs plus built-in cellular communications capability Mk1D will replace the Mk1C units.
Parthian Shots
There's a bunch of interesting platform evolution here. ONT simplifying their lineup in a number of places - focusing on a single pore chemistry, a single compute tower for PromethIONs - should pay dividends both from reducing customer confusion as well as ameliorating ONT's often spotty performance on actually delivering products and supporting them. If you have only one flowcell type, you can't ship the wrong ones! But, it's a small stream of simplification against a tide of further platform complexity.
The combination of Short Fragment Mode, PromethION P2 and Bonito makes for an interesting offering in the short read space. It would seem like an easy sell -- for the fraction of the cost of a desktop optical sequencer you can get similar amounts of data per run at a similar consumable cost. Of course, even with ONT's rosy accuracy claims the downstream analytic pipeline will be different. On yet the other hand, you can get methylation. It will be interesting to watch whether ONT can actually break into this market.
A few items made it only on the final release schedule slide covering into early June. One such "barely mentioned" item the long-run "Marathon" cells for PromethION weren't verbalized but early access is listed on the release date slide. The slide, shown below, is a tad confusing, with Kit 14 releases prior to R10.4.1 release -- Kit 14 contains R10.4.1. There's also the enigmatic "Kit 14 Amplification Range" - I don't remember that workflow branding before and a quick search of the Nanopore Community didn't refresh my memory (and no, I won't be surprised if it's painfully obvious or even mentioned in a prior blog post -- though Google seems to be making the latter unlikely)
If I were really clever, I'd have invested time reviewing my prior reports and noting what wasn't talked about in this presentation nor showed up on the release slide. Clive did say some announcements have been held back for London Calling, but it's still potentially interesting what has disappeared. VolTRAX was barely mentioned in the talk, but the few mentions underscored it continues as part of the platform. There are some items that simply make sense to expire quietly: with Bonito and duplex sequencing pushing accuracy, schemes such as doping with base analogs slide down the "making sense" scale. But others bear watching for at LC. For example, a new rapid chemistry using Vaccinia recombinase had been described that had the wonderful property of preserving input fragment lengths rather than shearing them as a transposase does. Still a project or crashed and burned?








2 comments:
Are these "Q's" frequently mentioned by ONT just Phred scores in FASTQ files? If so, they account only for point sequencing errors and not for errors that result in deletion and insertion of bases, right?
Also MIA were voltage sensing, new small diameter wells, the supposedly better "silver bullet" chemistry and any mention of the new chip designs they teased a tear or two ago.
Post a Comment