A story: back around the time Element launched AVITI, and Singular was mired in their hardware supply chain issues, a longtime friend / former colleague met me for breakfast and mentioned that he had bought his startup a MiSeq. I loved that they had bought a sequencer and thought MiSeq wwouas ideal for the company's immediate goals, but commented that if he had consulted me I would have urged that they consider AVITI. It would have been a bit of risk -- the platform was still new - and a bit pricey - about 3X a MiSeq - but AVITI would cover their future plans. My optimism seems to have been well-placed; Element has continued to expand the functionality and ease-of-use of the platform.
Something users asked for that wasn't on Element's roadmap was a way to load the lanes on the flowcell independently; Element was able to adapt their library loading fixture to have an auxiliary position to accommodate this. Contrast this with Illumina instruments which require the user to purchase a tool and manually load each lane.
Element also released medium and low output kits for the instrument, so that users now can choose 2x150 at 1B (High), 500M (Medium) or 125M (Low) clusters, with the 250M at about one half the price of the 1B - so the cost per gigabase is best with higher output, but users not requiring so much data can save money. The smaller kits also have shorter runtimes: it's 38 hours for the flagship 1B kit but 33 hours for 500M and 29 hours for 250 M.
Element is also addressing output and quality at both ends - there's a higher quality, fewer sequences UltraQ option giving Q50 quality and a lower quality, more sequences "Expert Mode HD" (HD=High Density).
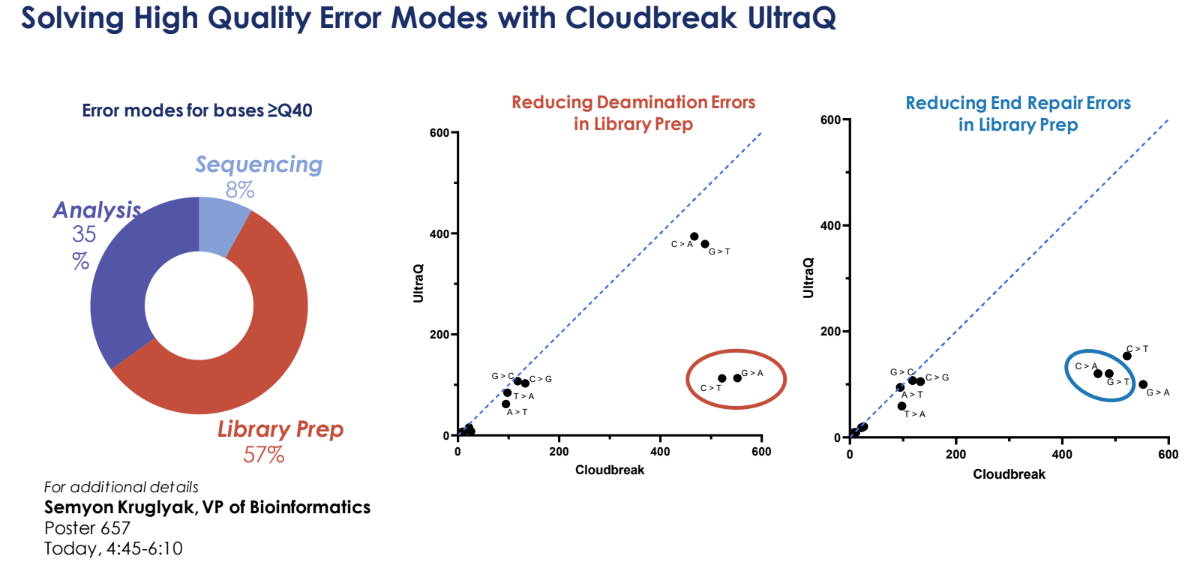
UltraQ follows the trend with PacBio Onso and now Ultima ppmSeq mode of delivering much higher quality than has been typical with short reads. As the other vendors have noted, this not only involves carefully tuning the chemistry and informatics, but also paying close attention to library prep methods. For example, one contributor to errors is deamination of cytosine to uracil, so the Q50 library prep scheme includes USER digestion to remove uracil.
Element also found that gap filling during blunting contributes a significant number of errors to the beginning of read 2 (a stunning dip in the accuracy plot below). Now, it might seem like just informatically trimming out those bases would work, but Element improves on that - they run 15 dark cycles at the beginning of Read 2, extending from the primer 15 bases but not probing with Avidites. This shows off the very low phasing of AVITI UltraQ chemistry - effectively their 2x150 is 1x150 1x165(forget the first 15) - the phasing rates have been reduced from 0.15% per cycle to only 0.08%. Pre-phasing (moving ahead a nucleotide) component is reduced from 0.04% to 0.01%. Read 2 has lower quality than Read 1 on UltraQ (with a curious climb in quality across Read 2), but both deliver Q50 for many positions. Element hopes to eliminate the dark cycles in the future by working with library prep partners to eliminate the errors at the start of Read 2. There's a nice talk from Element's Semyon Kruglyak available on their website that walks through this.
Expert mode isn't a different kit; it is simply an option within the operating software that tolerates higher cluster densities and hence a larger tail of lower quality sequences. The "Expert" part is this is intended for experts who understand the dependency of their application on quality scores and know that lower quality can be tolerated. In the 2x75 RNA-Seq example shown, Expert Mode yields about 30% more sequences.
Element over the past year released two different 2x300 kits - once an option available only on the venerable MiSeq and a favorite for certain applications such as 16S sequencing.
The biggest change for standard workflows is Cloudbreak Freestyle. AVITI uses circular library molecules whereas Illumina uses linear ones. So that Element users could access the vast Illumina-compatible ecosystem of library preparation methods, Element offered kits for converting libraries from linear to circular form. With Cloudbreak Freestyle, this conversion occurs on AVITI. Particularly for labs running a mix of Illumina and AVITI instruments, this takes an annoying detour out of the workflow when libraries are ultimately going on AVITI.
But, as the late night ads say, that's not all! Hybrid capture workflows are popular in many settings, but involve a number of molecular biology steps including multiple bead cleanups. Element's Trinity workflow, scheduled for release in Q3 of this year, has a single probe binding step off-instrument and then the remaining workflow steps occur on AVITI. That will be followed with Trinity PLus, which will perform all of the enrichment chemistry on AVITI. Element also showed data supporting that Trinity works just as well with 62.5ng of input DNA versus 500 nanograms -- and that's total load, after pooling multiple libraries -- enabling hybrid capture from more precious samples.
Yet in Element's tests Trinity performed as well or better than an unnamed conventional solution capture approach, with better %GC uniformity and library complexity and better variant calling - particularly at extremes of %GC. Trinity will allow exome capture, smaller targeted panels and custom panels - and even work with third-party capture reagents. By the way, I love how Element's Shawn Levy gave a historical overview of hybrid capture enrichment, reaching back to the early days of the Human Genome Project.
In one of the morning in-suite talks (again, lucky for me this is recorded as my breakfasts were all booked) PhaseGenomics CEO Ivan Liachko described yet another advantage of AVITI over Illumina: lower duplication rates. The patterned flowcells used on newer Illumina instruments have enabled very high cluster densities and a tolerance for a wider range of loading concentrations, but have brought two pain points from the Exclusion Amplification (ExAmp) method used to cluster within the nanowells of the patterned array. The most notorious nuisance of ExAmp is a very high index hopping rate, which I know from painful experience is extremely recalcitrant to any sort of attempt to experimentally eliminate. Trust me on that. But the less publicized issue very relevant to the OncoTerra proximity ligation libraries generated by PhaseGenomics, is that ExAmp generates higher duplicate rates. This is presumably due to amplified library molecules escaping one cluster and seeding another -- perhaps by tipping over into an adjacent well but I've also heard of analyses suggesting they can migrate over significant distances. Ivan commented that the tipping over may be more severe with proximity ligation libraries, as the inserts tend to be on the larger side of Illumina inserts - plus proximity ligation libraries are inherently less diverse than typical shotgun libraries. Element's rolling circle clustering has low index hopping and low duplication rates - even on OncoTerra libraries made from FFPE oncology samples. In the most extreme case, an OncoTerra library had nearly 30% duplication rate on NovaSeq 6000 which dropped to nearly zero on AVITI.
So where next? An obvious path would be a "big AVITI" that deliverers higher throughput. But so far, Element seems very focused on having a single instrument -- well, the AVITI24 to support Teton spatial profiling has upgraded compute, but is really still the same hardware to support chemistry. So if not new hardware, then what? Well, tuning a wide range of enrichment challenges for the on-instrument enrichment might keep them busy for a while. Could other library types benefit from on-instrument processing to simplify workflows? And since AVITI can support long inserts - perhaps up to a kilobase - and has very low phasing, perhaps kits offering longer than 2x300 might show up if customers demanded them.
[2024-02-29 12:26 -- fixed 1M to 1B for flagship flowcell ]






No comments:
Post a Comment