Before diving into today's presentation, a bit of useful background framing. Last May at London Calling, Brown opened the first day by wowing the crowd (and those of us tuning in via Twitter) with a dazzling list of major platform upgrades. These included a much faster sequencing speed ("fast mode"), less expensive chips that separated electronics from the wet components ("crumpet chips"), a big sibling for MinION in PromethION, increased numbers of pores per device, etc. Some of these were projected to arrive later that summer or in the fall. However, while the list of publications and presentations showcasing MinIONs continued to grow, none of these upgrades were seen, and indeed grumbling about delays in PromethION delivery were reported at AGBT.
Apparently Oxford Nanopore is one place that values the Loman on the totem pole, as Nick tweeted a hint he was playing with fast mode. His theatrical life of a salesman continued with his talk at AGBT, in which he mentioned (already leaked on Twitter) that he and Matt Loose had prepared sequencing libraries in his hotel room. But, neither of these improvements had moved on to the larger crowd. Then came last month's legal actions by Illumina. So, the somewhat unexpected announcement of an hour of Clive presenting, with an enigmatic title, provoked eager interest.
I've rearranged the presented items into thematic groups; if you watch the hangout the order will be different, plus I'm pretty much skipping the intro material Clive gave. NextgenSeek has Storified tweets, and David Eccles has left a detailed set of notes as a comment over at homolog.us (an odd place to leave them, given that the same site doubled down today on its prediction of Oxford's imminent demise [a bizarre bit of business dowsing, to say the least))
R9 Chemistry
Much of the announcements centered around a new chemistry, termed R9. Indeed, everything hangs to some degree on R9, though many of the changes could have been independent. Clive explained first the naming scheme: poRes have R-numbers, motor Enzymes have E-numbers and Membranes have M-numbers. So R9 is a new pore which replaces the earlier R7 chemistry currently in use (R6 was the first chemistry released; R8 apparently won't see light-of-day).
A striking announcement was for the often uber-secretive Oxford to announce the identity of the R9 pore: it is E.coli CsgG, a component in the protein secretion system which generates curli. A nonameric protein with 36 beta sheets, it is quite different in architecture to the MspA pore over which Illumina claiming ownership. While Clive never mentioned the suit or Illumina (nor any colorful euphemism for their rivals), this announcement clarifies that the U Washington patents licensed by Illumina should be no barrier to further development of the Oxford platform in the U.S.. Using CsgG for nanopore sensing is licensed from VIB in Ghent, Belgium. Brown said that Oxford had screened over 700 mutants of this pore, though the number of mutations in the R9 chemistry was not stated.
Furthermore, fast mode is now going to a reality: with the new kits (going to developers this month and the whole community next month, with R7 phased out shortly thereafter), speeds of 280 bases per second (or about 3-fold higher than the current 80bps). The system is believed to be capable of even faster; Oxford plans to roll out software updates to up the speed to closer to 500 bps (perhaps in steps) as they gain confidence in the new system.
New Basecaller
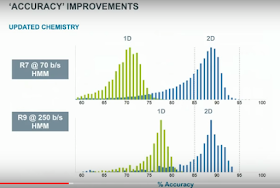
As Oxford has hinted before, the R9 chemistry in fast mode has an inherent advantage for sequencing, what Clive termed 'non-exponential' event length distribution. This cuts down the number of events which are too fast for the electronics to differentiate, which is believed to be a major source of base deletion errors. Simply running in fast mode both upshifts the accuracy curve and seems to tighten it up a bit!
An interesting difference in the R9/CsgG pore vs. the R7 pore (whose identity remains undisclosed) is that the signal is now mostly generated by only two contiguous bases, with a third base contributing significantly less -- but there are also "edge effects" from additional bases interacting with the pore.
In part due to this, Oxford is moving their basecalling to a new algorithm, Recurrent Neural Networks (RNNs), versus. the prior Hidden Markov Model scheme. These in turn utilize components called bidirectional Long Short Term Memory (BLSTM). I found a useful intro to these, at least useful for me, as I hadn't encountered these in bioinformatics before. A key difference over simpler neural network schemes (which long-ago as an undergraduate I got excited about) is that there are cycles in the graph structure for the network; nodes can feed back on themselves. That's all I've gotten so far; any pointers to other tutorials would be great to leave in the comments!
For users, the key bit is the RNN/BLSTM caller pushes the accuracy even further, and Brown was definitely of the opinion that further improvement should be achieved. Indeed, the ultimate goal is to have 1D reads with 90% accuracy and 2D reads with 99% accuracy.
In addition to a new algorithm, Oxford will release the source code for both this and the older 1D basecaller to parties who have signed their Developer's License (as Clive was quite frank about, Oxford is not interested in enabling potential competitors). Local versions of the 1D caller will be rolled out in a new MinKNOW, which has been a popular wishlist item for many users. Clive still thinks 2D calling belongs in the cloud -- it is both memory and compute intensive. Wider support for run-until will also be among these releases.
Clive somewhat obliquely hinted that fast mode will entail a change in the FAST5 files (or perhaps even a shift from FAST5), but no details were given.
1D MuA Library Prep
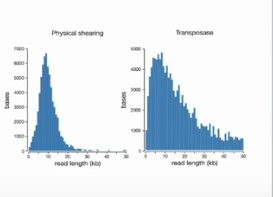
Nanopore has talked about a fast, easy library preparation method for a while, but now it will become a reality. A MuA transposase-base kit will enable making 1D libraries in 10 minutes and using only 2 tubes -- and I have it from Nick there are no bead cleanups. As Nick and Matt demonstrated at AGBT, this means sequencing with little more than pipettors -- if you have clean DNA. Clive mentioned that Oxford continues to work on their VolTRAX microfluidic device for sample-to-sequencer preparation, but no updates at this time -- wait for London Calling in May (a refrain during the talk).
Even more stunning than the idea of a 10 minute prep requiring only pipetting is that the size distribution for the reads looks better than results with G-tubes in terms of a tail of really long reads! Given that the accuracy of 1D is claimed to be approaching that of Pacific Biosciences, it seems plausible (but not yet proven) that these reads can feed into available assembly algorithms.
Clive was asked about short fragment size libraries with the MuA kit (e.g. for applications such as rapid aneuploidy detection); this is something they would need to try to tune. Clive also touched on size selecting fragments; he sees no reason this can't be done, but would prefer to tune the sample prep to obviate the need for it.
The MuA prep is doubly exciting for me. First, I'm all thumbs in the lab. For a long time, the only injury log entry at Starbase had my name on it -- if you haven't flame-sterilized labware in 2 decades, some of the subtle aspects of this operation slip your memory.. Second, with such a small investment in generating libraries, the number of opportunities for routine sequencing in a standard molecular biology lab are enlarged. Indeed, this is a powerful combination -- the training curve is flattened to nearly nothing and the effort required to sequence becomes minimal.
Blocking
An interesting side trip by Clive was the topic of blocking. This is his term for the loss of flowcell performance over time, which can steal as much as 50% of the theoretical yield of a flowcell. While he again put off details for London Calling, a tantalizing claim of finding a "smoking gun" was made, and it was suggested that nicked DNA and supercoiling may play roles. In the Q&A at the end, Clive opined that he believes G4 quartet structures are not an issue, which had been suggested in a pre-print from a MAP group.
PromethION
As noted above, reports have circulated that no PromethION devices had been shipped. Brown confirmed this, but stated that the first devices will ship at the end of the month. Oxford is assembling them at a pace of about 4-6 now and hoping to hit 10 per month by year's end (also clearing the queue by that time), with further scaling next year. He also offered that anyone who made a deposit and would like that refunded may request it, but they will lose their place in the queue -- deliveries are being made in the order deposits were made. One cause of delay has been a component (part of the physical connection between the electronics and the instrument, I think) being sent back to the manufacturer due to failing specifications.
Clive reviewed the basic specs of the machine, and explicitly compared its throughput to HiSeq X (with an ultimate goal of over 6 terabases per day!). The electronics (ASICS) can monitor 3K channels, with 144K channels across the array of 48 flowcells. Flowcells are completely independent of each other; the box can run a single flowcell or the array of 48 can be started in any crazy-quilt order you (or your sample flow) desires. Each flowcell has 4 independent compartments, which can be further multiplied by barcoding samples. Pores are R9 only; R9.2 initially though Clive hopes that R9.3 will be ready before flowcells ship. Input DNA of "less than 100ng" should result in enough libraries for 4 loadings, which could be 1 flowcell with 4 reloads, 2 flowcells with 2 reloads or 4 flowcells loaded once. Instrument geometry allows for loading with standard multichannel pipettors.
PromethION will come with serious compute on-board, allowing for local 1D basecalling: with 12 i7 quad-core nodes (later to optionally grow to 24) driven by the SLURM scheduling system. Clive envisions 2D moving to this cluster in the future (or just having 1D so good nobody bothers with 2D), and the architecture should allow very flexible application of the compute -- if you want to throw the entire cluster at calling Control is via a web interface, enabling even "a simple tablet" to control the device.
Odds and Ends
Just to round out, what were topics either sloughed off to London Calling or simply not mentioned?
Direct RNA sequencing came up in multiple questions; Clive said this is an area of active exploration but suggested that ONT still has not found a motor they are happy with for this application. He also thought that RNNs might be a better algorithm for RNA, though no elaboration was given. A related request came in for a combined RNA+DNA prep (wow, could that be killer for infectious disease detection!); Clive said he needed to think about that.
Protein analysis was also asked about; Brown sounded like this is still of interest but very back-burner. As Nick Loman has emphasized on Twitter, CsgG's natural role is in peptide translocation, so protein analysis is a tantalizing possibility.
Crumpet chips were not directly mentioned, but one question concerned "$20 flowcells" -- Clive essentially said wait until London Calling -- his belief is that the blocking issue is a higher priority.
No mention was made of increasing the number of pores per flowcell on MinION.
As noted before, disclosure further development of VolTRAX was also put off to London Calling.
One item Clive made clear is a non-topic is a MinION MkII -- all of the changes are in the consumables and software, with no plans for changing the MinION hardware.
Closing Thoughts
Oxford Nanopore made a lot of key announcements, making clear that the recent legal action will not have lasting effects on their platform, enabling very fast preps with very little expertise, and enhancing the throughput and speed (which, as I've emphasized before, are largely one-and-the-same). The status of PromethION was greatly clarified,
With these devices in the field, it will be interesting to see what splashy experiments are run prior to London Calling. I'm guessing that once burn-in is done, the first labs to get PromethIONs will be itching to throw human samples on them; the first human genome done on nanopore I expect will be out no later than the end of summer, and again I wouldn't be shocked for a rough one to be rammed out before London Calling (or a dataset slammed out by Oxford at London Calling).
Logistics and sticking to release schedules has not been a strong suit for Oxford; while this is unsurprising for a radical new technology, user base patience with this will be more difficult to find as the user base enlarges (or perhaps more accurately, the distribution of patience will broaden). Delivering these updates on time will win valuable credibility. Whether Oxford will be more cautious with updates at London Calling will be interesting to see; given user excitement over the platform in its current state, a more measured approach could help retain that enthusiasm.







G4 quartet structures......do you mean quadraplex?
ReplyDeleteSeems like the R9 2D reads have accuracy slightly better than PacBio by looking at the graph. PacBio can be in real trouble if this is true.
ReplyDeleteQuarte-tomato quadra-tomahto :-) - but you are right, Google finds an overwhelming preference for the term G4 quadruplex, so I shall endeavor to use that hencefourth.
ReplyDeleteyanks! always screwing with English language!
ReplyDelete"a comment over at homolog.us (an odd place to leave them)"
ReplyDeleteDavid had been commenting on nanopore technology for over a year. The technology is interesting, and David is very excited about it. Whenever he comments, we make it as the top on our blog and also notify twitter users.
http://www.homolog.us/blogs/blog/2014/10/19/nanopore-updates-from-david-eccles/
"given that the same site doubled down today on its prediction of Oxford's imminent demise"
Doubled down today? That is a bit dishonest. The prediction came before Illumina's patent lawsuit/FTC filing, and we had no insider information about the lawsuit.
"a bizarre bit of business dowsing, to say the least"
So, I guess the blogs are supposed to be only for pumping up tech companies !! Do you think there could be another side, who may be helped by accurate information or like to hear both pluses and minuses?
Yes, we are pessimistic about the business because of its valuation. All interesting technologies do not make multi-billion dollar businesses.
"An interesting difference in the R9/CsgG pore vs. the R7 pore (whose identity remains undisclosed)"
ReplyDeleteAny speculation on the identity of R7?
Why do people still believe anything that comes out of Clive's mouth? It's been over 4 years since AGBT 2012 and how many "announcements" made from Clive then came true? This guy has a horrible track record of BS. He's an academic, not an engineer and clearly has no experience in going from research to commercialization. I mean common, the system was delayed because of a out of spec connection component? REALLY!? It's also real easy to show accuracy plots with the mean <80% but say your long term goal is above 90%. Obviously your long term goal is going to be high. But why has it been 2 years and 1D accuracy is still in the 70% range? At this rate it will take 6 years to get it to the 90's if its even possible.
ReplyDeleteIt's clearly close to fund raising/ipo time for oxford so Clive is at it again.
"Why do people still believe anything..."
ReplyDeletemaybe because they got a MinION ? I went to look at their AGBT12 slides and it seems to me they've done 8 or 9/10 of the things they said certainly on MinION.
1. Portable USB DNA sequencer. Yes.
2. Less than $1000. Yes
3. > 500MB in 6 hrs. Yes.
4. Ultra long reads. Yes (given the right preps).
5. Robust membranes. Yes.
6. 10 minute sample preps. Yes
7. Open software. Yes (recently).
8. Base modifications. Yes.
9. Assemblies/ Yes
Whats left is error rate at < 4% according to the slides. That isnt there yet but this talk indicates its not far off, as is much higher throughput and lower costs than they originally promised. Although i note they do go on about the ASIC and the enzyme speed and room for improvement. I was at AGBT when PacBio promised a 15 minute $1000 genome within a couple of years ! PacB are doing just fine too.